| J Korean Diabetes > Volume 21(3); 2020 > Article |
|
Abstract
Recently, machine learning (ML) applications have received attention in diabetes and metabolism research. This review briefly provides the basic concepts of ML and specific topics in diabetes research. Exemplary studies are reviewed to provide an overview of the methodology, main findings, limitations, and future research directions for ML-based studies. Well-defined, testable study hypotheses that stem from unmet clinical needs are always the first prerequisite for successful deployment of an MLbased approach to clinical scene. The management of data quality with enough quantity and active collaboration with ML engineers can enhance the ML development process. The interpretable highperformance ML models beyond the black-box nature of some ML principles can be one of the future goals expected by ML and artificial intelligence in the diabetes research and clinical practice settings that is beyond hype. Most importantly, endocrinologists should play a central role as domain experts who have clinical expertise and scientific rigor, for properly generating, refining, analyzing, and interpreting data by successfully integrating ML models into clinical research.
ļ©ĖņŗĀļ¤¼ļŗØ(machine learning, ML)ņØĆ ņĄ£ĻĘ╝ ļ│æņøÉ ņ×ÉļŻīņØś ņĀäņé░ĒÖö, ņ×äņāüņ×ÉļŻī ļłäņĀü, ļīĆĻĘ£ļ¬© ņ×ÉļŻīļź╝ ņ▓śļ”¼ĒĢĀ ņłś ņ׳ļŖö ņ╗┤Ēō©Ēä░ņØś ņĀĆņן ļ░Å Ļ│äņé░ļŖźļĀźņØś ņ”ØĻ░ĆņÖĆ ĒĢ©Ļ╗ś ļ¦ÄņØĆ Ļ┤Ćņŗ¼ņØä ļ░øņ£╝ļ®░ ņŚ¼ļ¤¼ ņ×äņāü ļČäņĢ╝ ņŚ░ĻĄ¼ņÖĆ ņ¦äļŻīņŚÉņä£ ĒÖ£ņÜ®ļÉśĻ│Ā ņ׳ļŗż. ļŗ╣ļć©ļ│æĻ│╝ ļé┤ļČäļ╣äņ¦łĒÖś ļČäņĢ╝ņŚÉņä£, ļ©ĖņŗĀļ¤¼ļŗØĻ│╝ ņŚ░Ļ┤ĆļÉśņ¢┤ Ļ▓īņ×¼ļÉ£ ļģ╝ļ¼ĖņØś ņłśļŖö PubMed Ļ▓ĆņāēĻĖ░ņżĆ 1986ļģäļČĆĒä░ 2020ļģä 1ņøöĻ╣īņ¦Ć ņĢĮ 2,000Ļ▒┤ņŚÉ ļŗ¼ĒĢśļ®░, ņĄ£ĻĘ╝ 10ļģäĻ░ä ĻĖ░ĒĢśĻĖēņłśņĀüņØĖ ņ”ØĻ░ĆņČöņäĖļź╝ ļ│┤ņØ┤Ļ│Ā ņ׳ļŗż. ļ│Ė ņóģņäżņŚÉņä£ļŖö ļ©ĖņŗĀļ¤¼ļŗØĻ│╝ ņŚ░Ļ┤ĆļÉ£ ņŚ¼ļ¤¼ Ļ░£ļģÉņŚÉ ļīĆĒĢ£ Ļ░äļץĒĢ£ ņĀĢņØś, ļŗ╣ļć©ļ│æĻ│╝ ļé┤ļČäļ╣äņ¦łĒÖśņŚÉņä£ ļ©ĖņŗĀļ¤¼ļŗØņØä ĒÖ£ņÜ®ĒĢ£ ņśłņŗ£ ņŚ░ĻĄ¼ļōżņØä ņé┤ĒÄ┤ļ│┤Ļ│Ā, ņČöĒøä ļŗ╣ļć©ļ│æ ļ░Å ļé┤ļČäļ╣äņ¦łĒÖś ņ×äņāüņŚ░ĻĄ¼ņÖĆ ņ¦äļŻīņŚÉ ņ׳ņ¢┤ ļ©ĖņŗĀļ¤¼ļŗØņØś ĒÖ£ņÜ®ļ░®ņĢł ļ░Å ļ░£ņĀäĻ░ĆļŖźņä▒ņŚÉ ļīĆĒĢ┤ ļģ╝ņØśĒĢśĻ│Āņ×É ĒĢ£ļŗż.
ņØĖĻ│Ąņ¦ĆļŖź(artificial intelligence, AI), ļ©ĖņŗĀļ¤¼ļŗØ(ML), ļöźļ¤¼ļŗØ(deep learning)ņØĆ ņØśĒĢÖ ļČäņĢ╝ņŚÉņä£ ļ¦ÄņØĆ Ļ▓ĮņÜ░ ļÅÖņŗ£ņŚÉ ņé¼ņÜ®ļÉśĻ▒░ļéś Ļ░ÖņØĆ ņØśļ»ĖļĪ£ Ēś╝ņÜ®ļÉśĻ│Ā ņ׳ļŗż. ļ»ĖĻĄŁņŗØĒÆłņØśņĢĮĻĄŁ(U.S. Food and Drug Administration)ņØĆ ņØĖĻ│Ąņ¦ĆļŖźņØä ņĪ┤ļ¦źņ╗żņŗ£ņØś ņĀĢņØśņŚÉ ļö░ļØ╝ ŌĆśņ¦ĆļŖźņØ┤ ņ׳ļŖö ĻĖ░Ļ│äļź╝ ļ¦īļō£ļŖö Ļ│╝ĒĢÖĻ│╝ Ļ│ĄĒĢÖ(the science and engineering of making intelligent machines, especially intelligent computer programs)ŌĆÖņ£╝ļĪ£ ņĀĢņØśĒĢśņśĆļŗż[1,2]. ņØ╝ļ░śņĀüņ£╝ļĪ£ ņ¦ĆļŖźņØĆ ņ×ÉĻĖ░ņØĖņŗØ, Ļ▓ĮĒŚśņĀü ņ¦ĆņŗØ ļō▒ ĒżĻ┤äņĀüņØĖ ņśüņŚŁņØä ņ¦Ćņ╣ŁĒĢśļŖö Ļ░£ļģÉņØ┤ņ¦Ćļ¦ī, ĻĘĖ ņżæ ĒĢÖņŖĄĻ│╝ ņČöļĪĀņØĆ Ēśä ņŗ£ņĀÉņŚÉņä£ ņØĖĻ│Ąņ¦ĆļŖźņØä ņĀĢņØśĒĢśļŖö Ļ░Ćņן ņżæņÜöĒĢ£ ņÜöņåīļĪ£ ņØĖņ¦ĆļÉśĻ│Ā ņ׳ļŗż[1]. ņØĖĻ│Ąņ¦ĆļŖźņØ┤ Ļ░Ćņן ĒżĻ┤äņĀüņØĖ ņāüņ£ä Ļ░£ļģÉņØ┤ļØ╝ļ®┤, ļ©ĖņŗĀļ¤¼ļŗØņØĆ ŌĆśļŹ░ņØ┤Ēä░ļĪ£ļČĆĒä░ ņŖżņŖżļĪ£ ĒĢÖņŖĄĒĢśņŚ¼ ļČäļźśļéś ņśłņĖĪ ļō▒ņØś ņłśĒ¢ēļŖźļĀźņØä Ļ░£ņäĀĒĢĀ ņłś ņ׳ļŖö ņåīĒöäĒŖĖņø©ņ¢┤ļéś ņĢīĻ│Āļ”¼ņ”śņØä ļööņ×ÉņØĖŌĆÖĒĢśĻĖ░ ņ£äĒĢ£ ņØĖĻ│Ąņ¦ĆļŖźņØś ĒĢ£ ļČäņĢ╝ļĪ£ ņĀĢņØśļÉĀ ņłś ņ׳ļŗż. ļö░ļØ╝ņä£ ļ©ĖņŗĀļ¤¼ļŗØņØĆ ņØĖĻ│Ąņ¦ĆļŖźņŚÉ ĒżĒĢ©ļÉśļŖö Ļ░£ļģÉņØ┤ļéś, ņØĖĻ│Ąņ¦ĆļŖźņØ┤ Ļ╝Ł ļ©ĖņŗĀļ¤¼ļŗØņØä ņ¦Ćņ╣ŁĒĢśļŖö Ļ▓āņØĆ ņĢäļŗłļŗż. ļöźļ¤¼ļŗØ Ēś╣ņØĆ ņŗ¼ņĖĄņŗĀĻ▓Įļ¦Ø(deep neural network)ņØĆ ļ©ĖņŗĀļ¤¼ļŗØņØś ĒĢ£ ņóģļźśļĪ£, ņŚ¼ļ¤¼ Ļ▓╣ņØś ņŗĀĻ▓Įļ¦ØņØä ņīōņĢä ņé¼ļ×īņØś ļæÉļćīņÖĆ ņ£Āņé¼ĒĢśĻ▓ī ņäżĻ│äĒĢ£ ņĢīĻ│Āļ”¼ņ”śņØä ņ¦Ćņ╣ŁĒĢ£ļŗż. 2018ļģä Ļ░£ņĄ£ļÉ£ 3ņ░© ML for Health workshopņŚÉ ļö░ļź┤ļ®┤, ņĀäĒåĄņĀüņØĖ Ļ│ĄĒĢÖļČäņĢ╝ ņŚöņ¦Ćļŗłņ¢┤ ļ░Å ņŚ░ĻĄ¼ņ×ÉļōżņØĆ ļ│ĖņØĖļōżņØś ņŚ░ĻĄ¼ļČäņĢ╝ļź╝ ņ¦Ćņ╣ŁĒĢĀ ļĢī ļ©ĖņŗĀļ¤¼ļŗØņØ┤ļØ╝ļŖö ļŗ©ņ¢┤ļź╝ ļŹö ņäĀĒśĖĒĢśņśĆņ£╝ļéś, ņ×äņāüņØśņé¼ļōżņØĆ ļŹö ļäōņØĆ ņØśļ»ĖņØś ņØĖĻ│Ąņ¦ĆļŖźņØ┤ļØ╝ļŖö ļŗ©ņ¢┤ļź╝ ĒØöĒ׳ ĒÖ£ņÜ®ĒĢśņśĆļŗż. ņŚ░ĻĄ¼ ņ╗żļ«żļŗłĒŗ░ Ļ░ä ņÜ®ņ¢┤ĒÖ£ņÜ® ņ░©ņØ┤ļŖö ņåīĒåĄ ņןņĢĀ ļ░Å ļČłĒĢäņÜöĒĢ£ ņśżļźśļź╝ ņ£Āļ░£ĒĢĀ Ļ░ĆļŖźņä▒ņØ┤ ņ׳ņ¢┤, ļ©ĖņŗĀļ¤¼ļŗØ Ļ┤ĆļĀ© ņÜ®ņ¢┤ņØś ņĀĢĒÖĢĒĢ£ ņĀĢņØś ļ░Å ņé¼ņÜ®ņŚÉ ļīĆĒĢ£ Ļ│ĄĒåĄņØś ļģ╝ņØśĻ░Ć ņČöĒøä ĒĢäņÜöĒĢĀ Ļ▓āņ£╝ļĪ£ ļ│┤ņØĖļŗż[3].
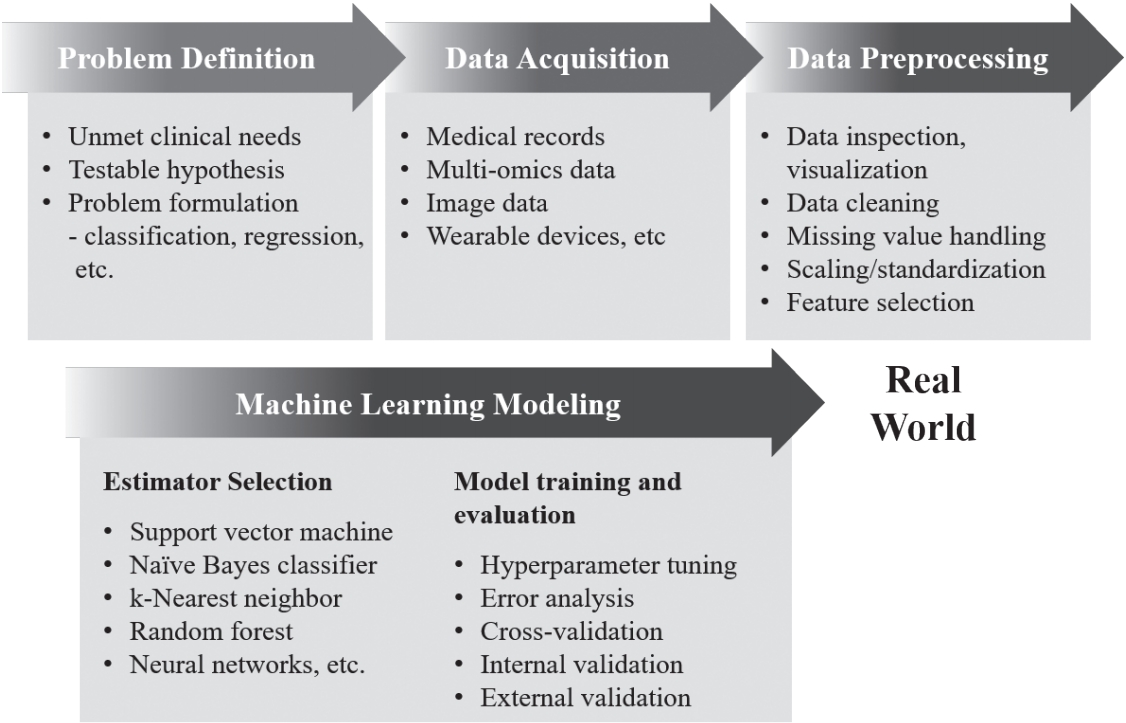
ļ©ĖņŗĀļ¤¼ļŗØ ņĢīĻ│Āļ”¼ņ”śņØĆ Ēü¼Ļ▓ī ņ¦ĆļÅäĒĢÖņŖĄ(supervised learning), ļ╣äņ¦ĆļÅäĒĢÖņŖĄ(unsupervised learning), ņżĆņ¦ĆļÅäĒĢÖņŖĄ(semi-supervised learning), Ļ░ĢĒÖöĒĢÖņŖĄ(reinforcement learning) 4Ļ░Ćņ¦Ć ļ▓öņŻ╝ļĪ£ ļéśļłäņ¢┤ ļ│╝ ņłś ņ׳ļŗż(Table 1) [4-7]. ņ¦ĆļÅäĒĢÖņŖĄņØĆ ļ©ĖņŗĀļ¤¼ļŗØ ņĢīĻ│Āļ”¼ņ”śņØä ĒĢÖņŖĄņŗ£ĒéżĻĖ░ ņ£äĒĢ┤ ļŹ░ņØ┤Ēä░ņÖĆ ņĀĢļŗĄņ¦Ć(ļĀłņØ┤ļĖö, label)ļź╝ ĒĢ©Ļ╗ś ĒĢäņÜöļĪ£ ĒĢ£ļŗż. ņ¦ĆļÅäĒĢÖņŖĄņØś ļ¬®Ēæ£ļŖö ĻĖ░ņĪ┤ ļŹ░ņØ┤Ēä░ņÖĆ ļĀłņØ┤ļĖöĻ│╝ņØś Ļ┤ĆĻ│äļź╝ ļ░öĒāĢņ£╝ļĪ£, ņāłļĪ£ņÜ┤ ļŹ░ņØ┤Ēä░Ļ░Ć ļōżņ¢┤ņÖöņØä ļĢī ĻĖ░ņĪ┤ ņ×ÉļŻīņŚÉ ļ╣äņČöņ¢┤ Ļ░Ćņן ņĀüĒĢ®ĒĢ£ ņśłņĖĪĻ░ÆņØä ņČöļĪĀĒĢśļŖö Ļ▓āņØ┤ļŗż. ļ╣äņ¦ĆļÅäĒĢÖņŖĄņØĆ ļĀłņØ┤ļĖöņØ┤ ņŚåļŖö ļŹ░ņØ┤Ēä░ļź╝ ĒāÉņāēĒĢśļ®░, ĻĄ¼ņĪ░ļéś Ēī©Ēä┤ņØä ņ░ŠĻ▒░ļéś ņ░©ņøÉņØä ņČĢņåīĒĢśļŖö(ņĀĢļ│┤ļ¤ēņØ┤ ņāüļīĆņĀüņ£╝ļĪ£ ņĀüņØĆ ļ│ĆņłśļōżņØä ļ░░ņĀ£ĒĢśĻ│Ā ņĀäņ▓┤ ļŹ░ņØ┤Ēä░ļź╝ ņל ņäżļ¬ģĒĢśļŖö ņØ╝ļČĆ ļ│Ćņłśļōżļ¦ī ļé©ĻĖ░Ļ▒░ļéś ņäżļ¬ģļĀźņØ┤ ļåÆņØĆ ļ│Ćņłśļź╝ ņāłļĪ£ ņāØņä▒ĒĢśņŚ¼ ņĀäņ▓┤ ļ│Ćņłś ņłśļź╝ ņżäņØ┤ļŖö) ņ×æņŚģņØä ņłśĒ¢ēĒĢ£ļŗż. ņżĆņ¦ĆļÅäĒĢÖņŖĄņØĆ ņ¦ĆļÅäĒĢÖņŖĄĻ│╝ ļ╣äņ¦ĆļÅäĒĢÖņŖĄļ░®ņŗØņØä ĒĢ©Ļ╗ś ĒÖ£ņÜ®ĒĢśņŚ¼, ļīĆĻĘ£ļ¬© ļŹ░ņØ┤Ēä░ņŚÉņä£ ņØ╝ļČĆ ļŹ░ņØ┤Ēä░ņŚÉļ¦ī ļĀłņØ┤ļĖöņØ┤ ņĀ£ņŗ£ļÉśņ¢┤ ņ׳ņØä Ļ▓ĮņÜ░ ņ£ĀņÜ®ĒĢĀ ņłś ņ׳ļŗż. Ļ░ĢĒÖöĒĢÖņŖĄņØĆ ņŻ╝ņ¢┤ņ¦ä ļ│Ąņ×ĪĒĢ£ ĒÖśĻ▓ĮņŚÉ ļīĆĒĢ┤ ĒŖ╣ņĀĢ Ē¢ēļÅÖņØä ņĘ©Ē¢łņØä ļĢī ņŻ╝ņ¢┤ņ¦ĆļŖö ļ│┤ņāü Ēś╣ņØĆ ņåÉņŗżņŚÉ ļīĆĒĢ£ ĒĢÖņŖĄņØä ĒåĄĒĢ┤, ļłäņĀüļ│┤ņāüņØä ņĄ£ļīĆĒÖöĒĢśļŖö ņĄ£ņĀüņØś Ē¢ēļÅÖņØä ĒāÉņāēĒĢśļ®░, ņØ┤ļ¤¼ĒĢ£ ĒĢÖņŖĄņØĆ ņŗżņĀ£ ņ×æņŚģņØä ņ¦äĒ¢ēĒĢśļ®░ ļ░£ņāØĒĢśļŖö Ē¢ēļÅÖ-ļ░śņØæ ļŹ░ņØ┤Ēä░ļź╝ ĒåĄĒĢ┤ ņŗżņŗ£Ļ░äņ£╝ļĪ£ ņØ┤ļŻ©ņ¢┤ņ¦äļŗż. ĒĢ┤Ļ▓░ĒĢśĻ│Āņ×É ĒĢśļŖö ļ¼ĖņĀ£ņŚÉ ļīĆĒĢ┤ ņ¢┤ļ¢ż ļ©ĖņŗĀļ¤¼ļŗØ ņĢīĻ│Āļ”¼ņ”śņØä ņäĀĒāØĒĢĀ Ļ▓āņØĖĻ░ĆņŚÉ ļīĆĒĢ┤ ņØ╝ļČĆ ņ░ĖĻ│Ā Ļ░ĆļŖźĒĢ£ ņ¦Ćņ╣©ļōżņØ┤ ņ׳ĻĖ┤ ĒĢśņ¦Ćļ¦ī(ņśłņŗ£: ŌĆ£cheat-sheetŌĆØ for initial ML estimators, scikitlearn; https://scikit-learn.org/stable/tutorial/machine_ learning_map/index.html), ļīĆļČĆļČäņØś Ļ▓ĮņÜ░ ņĀüņĀłĒĢ£ ļ©ĖņŗĀļ¤¼ļŗØ ņĢīĻ│Āļ”¼ņ”śņØś ņäĀĒāØņØĆ ļŹ░ņØ┤Ēä░ņØś ņ¢æ, ĻĄ¼ņĪ░, ļ¼ĖņĀ£ Ēś╣ņØĆ Ļ░ĆņäżņØś ĒŖ╣ņä▒, ĻĖ░ņĪ┤ ņ¦ĆņŗØ, ņŚ░ĻĄ¼ņ×ÉņØś Ļ▓ĮĒŚśĻ│╝ ņ¦üĻ┤Ć, ĒĢÖņŖĄļÉ£ ņĢīĻ│Āļ”¼ņ”śņØś ņä▒ļŖź ļō▒ņØä Ļ│ĀļĀżĒĢśņŚ¼ ņ×ÉļŻīļź╝ ĒāÉņāēĒĢśĻ│Ā ļ¬©ļŹĖņØä ĻĄ¼ņČĢĒĢśļŖö ļ░śļ│ĄņĀüņØĖ Ļ│╝ņĀĢņØä ĒåĄĒĢ┤ ņØ┤ļŻ©ņ¢┤ņ¦äļŗż(Fig. 1). ņØ┤ļź╝ ņ£äĒĢ┤ ĒĢ┤Ļ▓░ĒĢśĻ│Āņ×É ĒĢśļŖö ļ¼ĖņĀ£ ļ░Å ļŹ░ņØ┤Ēä░ņØś ĒŖ╣ņä▒ņŚÉ ļö░ļØ╝ ļ©ĖņŗĀļ¤¼ļŗØ ļ¬©ļŹĖņŚÉ ļīĆĒĢ£ ņĄ£ņĀü ĒÅēĻ░Ćņ¦ĆĒæ£Ļ░Ć ņŗĀņżæĒ׳ Ļ▓░ņĀĢļÉśņ¢┤ņĢ╝ ĒĢ£ļŗż(Table 2) [8-10].
ņĄ£ĻĘ╝ 5ļģäĻ░ä 611Ļ░£ ņśüļ¼Ė ļģ╝ļ¼Ė(ņØĖĻ░ä ļīĆņāü, ņøÉņĀĆļ¦ī ĒżĒĢ©)ņØś ņĀ£ļ¬®ņØä ļČäņäØĒĢśņŚ¼ ņĀäņ▓┤ 2,155 ļŗ©ņ¢┤ ņżæ Ļ░Ćņן ļ╣łļ▓łĒĢśĻ▓ī ņé¼ņÜ®ļÉ£ 30Ļ░£ ļŗ©ņ¢┤ļź╝ ņČöņČ£ĒĢśņśĆņØä ļĢī, ņ¦łĒÖś ņżæņŚÉņä£ļŖö diabetesļéś diabeticņØ┤ Ļ░Ćņן ĒØöĒĢśĻ▓ī ļō▒ņןĒĢśņśĆņ£╝ļ®░(52%), retinopathy (14%), thyroid (14%)Ļ░Ć ļÆżļź╝ ņØ┤ņŚłļŗż. ļ©ĖņŗĀļ¤¼ļŗØ ļ¬®ņĀüņ£╝ļĪ£ļŖö detection, classification, identification, diagnosis ļō▒ ņ¦äļŗ© Ļ┤ĆļĀ© ļŗ©ņ¢┤Ļ░Ć 40%ļĪ£ Ļ░Ćņן ļ¦ÄņĢśņ£╝ļ®░, risk prediction Ēś╣ņØĆ predictionņØ┤ 31%ļĪ£ ņ░©ņł£ņ£ä, ĻĘĖ ņÖĖ segmentation (5%), Ēś╣ņØĆ bioinformatics (7%) ļō▒ņØ┤ ņ׳ņŚłļŗż. ņØ┤ ņżæ 1) ņ¦ĆļÅä, ļ╣äņ¦ĆļÅä, ņżĆņ¦ĆļÅä, Ēś╣ņØĆ Ļ░ĢĒÖöĒĢÖņŖĄ ĒÖ£ņÜ® ņŗżļĪĆļź╝ ņĀ£Ļ│ĄĒĢĀ ņłś ņ׳ņ£╝ļ®░, 2) ņĄ£ĻĘ╝ 3ļģä ņØ┤ļé┤ ļ░£Ēæ£ļÉ£ ļŗ╣ļć©ļ│æ ņŚ░Ļ┤ĆļČäņĢ╝ ņŚ░ĻĄ¼ 6ĒÄĖņØä Ļ▓ĆĒåĀĒĢśņśĆļŗż. ĒĢ┤ļŗ╣ ņŚ░ĻĄ¼ ņäĖļČĆ ļé┤ņÜ®ņØĆ Table 3ņŚÉ ņĀ£ņŗ£ĒĢśņśĆļŗż[4,5,11-14].
ļ¦ÄņØĆ ņŚ░ĻĄ¼ņ×ÉļōżņØ┤ ļ©ĖņŗĀļ¤¼ļŗØ ņĢīĻ│Āļ”¼ņ”śņØś ĒÖ£ņÜ®ņØ┤ ļŗ╣ļć©ļ│æ ļ░Å ļé┤ļČäļ╣äņ¦łĒÖśņØś ņäĀļ│äĻ▓Ćņé¼ ņä▒ļŖźĻ░£ņäĀņŚÉ ļÅäņøĆņØä ņżä ņłś ņ׳ņØäņ¦Ć ņŚ░ĻĄ¼ĒĢ┤ņÖöļŗż. Artzi ļō▒[4]ņØĆ ņĀäņ×ÉņØśļ¼┤ĻĖ░ļĪØ EHR ļŹ░ņØ┤Ēä░ļ▓ĀņØ┤ņŖżļź╝ ĒåĀļīĆļĪ£ ņ×äņŗĀņä▒ļŗ╣ļć©ļ│æņØä ņäĀļ│äĻ▓Ćņé¼ĒĢśļŖö ņĢīĻ│Āļ”¼ņ”śņØä Ļ░£ļ░£ĒĢśņśĆļŗż. 2010ļģäļČĆĒä░ 2017ļģäļÅäĻ╣īņ¦Ć ņØ┤ņŖżļØ╝ņŚś 36ļ¦īņŚ¼ ļ¬ģņØś ņ×äņé░ļČĆļĪ£ļČĆĒä░ ņłśņ¦æļÉ£ 58ļ¦īĻ▒┤ņØś EHR ņ×ÉļŻīļź╝ ĒÖ£ņÜ®ĒĢśņŚ¼ ņ×äņŗĀņä▒ļŗ╣ļć©ļ│æ ņäĀļ│äĻ▓Ćņé¼ ļ©ĖņŗĀļ¤¼ļŗØ ņĢīĻ│Āļ”¼ņ”śņØä ĒĢÖņŖĄņŗ£ņ╝░ļŗż. 2,355Ļ░£ņØś Ēøäļ│┤ ļ│Ćņłś ņżæ, ņŚ░ĻĄ¼ņ×ÉļōżņØĆ gradient boosting model ĻĖ░ļ░śņ£╝ļĪ£ 9Ļ░£ņØś ņ×ÉĻ░ĆņØæļŗĄņØ┤ Ļ░ĆļŖźĒĢ£ ļ│Ćņłśļ¦īņØä ĒÖ£ņÜ®ĒĢ£ ļ¬©ļŹĖņØä ļ¦īļōżņŚłĻ│Ā ņØ┤ļŖö ĻĖ░ņĪ┤ ņäĀļ│äĻ▓Ćņé¼ ļ░®ļ▓ĢņØĖ 24~28ņŻ╝ ņé¼ņØ┤ ļŗ╣ļČĆĒĢśĻ▓Ćņé¼ņŚÉ ļ╣äĒĢ┤ ļŹö ņĪ░ĻĖ░ņŚÉ ņŗ£Ē¢ēĒĢĀ ņłś ņ׳ņ£╝ļ®┤ņä£ļÅä ņóŗņØĆ ņä▒ļŖźņØä ļ│┤ņŚ¼ņŻ╝ņŚłļŗż(area under the receiver operating characteristics curve, 0.80 vs. 0.68).
ņל ĻĄ¼ņČĢļÉśĻ│Ā Ļ▓Ćņ”ØļÉ£, ņĀĢĒÖĢĒĢ£ ļ©ĖņŗĀļ¤¼ļŗØ ļ¬©ļŹĖņØĆ ĻĖ░ņĪ┤ņØś ņ╣©ņŖĄņĀü Ļ▓Ćņé¼ļź╝ ļīĆņ▓┤ĒĢĀ ņłś ņ׳ļŖö Ļ░ĆļŖźņä▒ņØ┤ ņ׳ļŗż. ņØ╝ļĪĆļĪ£ ļ╣äņĢīņĮ£ņä▒ņ¦Ćļ░®Ļ░ä(non-alcoholic fatty liver disease, NAFLD)ņØĆ ņĀäņäĖĻ│äņĀüņ£╝ļĪ£ ĻĖēņåŹĒ׳ ņ£Āļ│æļźĀņØ┤ ņ”ØĻ░ĆĒĢśĻ│Ā ņ׳ļŖö ņ¦łĒÖśņØ┤ņ¦Ćļ¦ī, ņŚ¼ņĀäĒ׳ ņĪ░ņ¦üņāØĻ▓ĆņØ┤ ĒÖĢņ¦äņØś gold standardļĪ£ ļÉśņ¢┤ ņ׳ņ¢┤ ņ¦äļŗ©Ļ│╝ņĀĢņØś ļČĆļŗ┤ņØä ņ┤łļלĒĢ£ļŗż. ĒĢ£ ņŚ░ĻĄ¼ņŚÉņä£ Ēśłņ▓Łņ£╝ļĪ£ļČĆĒä░ ļČäņäØĒĢ£ lipidomic, glycomic, liver fatty acid ļŹ░ņØ┤Ēä░ļź╝ ĒÖ£ņÜ®, support vector machine ĻĖ░ļ░ś NAFLD ņ¦äļŗ© ņĢīĻ│Āļ”¼ņ”śņØä ņĀ£ņŗ£ĒĢśņśĆļŗż[11]. Ļ░äņä¼ņ£ĀĒÖö ņĪ┤ņ×¼ ņŚ¼ļČĆņŚÉ ļīĆĒĢśņŚ¼, ĒĢ┤ļŗ╣ ļ¬©ļŹĖņØĆ 10Ļ░£ņØś ļŗ©ņł£ĒĢ£ lipid speciesļź╝ ĒÖ£ņÜ®ĒĢ£ ļ¬©ļŹĖļĪ£ļÅä 98%ņŚÉ ļŗ¼ĒĢśļŖö ņĀĢĒÖĢļÅäļź╝ ļ│┤ņŚ¼ņŻ╝ņ¢┤, lipidomicsņŚÉ ĻĖ░ļ░śĒĢ£ ļ©ĖņŗĀļ¤¼ļŗØ ņĢīĻ│Āļ”¼ņ”śņØ┤ Ļ░äņāØĻ▓ĆņŚÉ ļīĆĒĢ£ ļīĆņĢłņØ╝ ņłś ņ׳ļŖö Ļ░ĆļŖźņä▒ņØä ņĀ£ņŗ£ĒĢśņśĆļŗż. ļŗżļ¦ī, ņØ┤ļ¤¼ĒĢ£ ņĢīĻ│Āļ”¼ņ”śņØĆ Ļ░£ļ░£ņŚÉ ĒÖ£ņÜ®ļÉ£ ļŹ░ņØ┤Ēä░ņģŗņØś ĻĘ╝ļ│ĖņĀüņØĖ ņĪ░Ļ▒┤(ņØĖņóģ ļō▒)ņŚÉ ĻĄŁĒĢ£ļÉśņ¢┤ ļŗżļźĖ ņØĖĻĄ¼ņ¦æļŗ©ņŚÉņä£ņØś ņČöĻ░Ć Ļ▓Ćņ”ØņØ┤ ļ░śļō£ņŗ£ ĒĢäņÜöĒĢśļ®░ ĒĢ┤ļŗ╣ ņĢīĻ│Āļ”¼ņ”śņØś Ļ▓ĮņÜ░ Ļ▓Įņ”ØņØś NAFLDņŚÉ ļīĆĒĢ┤ņä£ļÅä ņóŗņØĆ ņä▒ļŖźņØä ĒÖĢļ│┤ĒĢĀ ņłś ņ׳ņØäņ¦ĆņŚÉ ļīĆĒĢ£ ņŚ░ĻĄ¼Ļ░Ć ņČöĻ░ĆļĪ£ ĒĢäņÜöĒĢśļŗż[15].
ņĀĢĒÖĢĒĢ£ ņ×äņāüĻ▓ĮĻ│╝ ņśłņĖĪņØĆ Ļ░£ļ│äĒÖöļÉ£ ņ╣śļŻī ļ░Å Ļ┤Ćļ”¼ļź╝ Ļ░ĆļŖźĒĢśĻ▓ī ĒĢ£ļŗżļŖö ņĀÉņŚÉņä£ ņżæņÜöĒĢśļŗż. WATCH-DM scoreļŖö ņĀ£2ĒśĢ ļŗ╣ļć©ļ│æ ĒÖśņ×ÉņŚÉņä£ ņŗ¼ļČĆņĀä ļ░£ņāØ ņ£äĒŚśļÅäļź╝ ņśłņĖĪĒĢśļŖö ļ©ĖņŗĀļ¤¼ļŗØ ļ¬©ļŹĖļĪ£, ĻĖ░ņĪ┤ ņל ņĢīļĀżņ¦ä ļ¼┤ņ×æņ£äļīĆņĪ░ĻĄ░ ņŚ░ĻĄ¼ņØĖ ACCORD ņŚ░ĻĄ¼ņ×ÉļŻīņŚÉņä£ ļ¬©ļŹĖņØä ĒĢÖņŖĄņŗ£ĒéżĻ│Ā ļśÉ ļŗżļźĖ ļīĆĻĘ£ļ¬© ļ¼┤ņ×æņ£äļīĆņĪ░ĻĄ░ ņŚ░ĻĄ¼ņ×ÉļŻīņØĖ ALLHAT ņŚ░ĻĄ¼ņ×ÉļŻīņŚÉņä£ ņÖĖļČĆĻ▓Ćņ”Ø(external validation)ņØä ņŗ£Ē¢ēĒĢśņŚ¼, ņŗĀļó░ĒĢĀļ¦īĒĢ£ ļ¬©ļŹĖņØś ņä▒ļŖźņØä ļ│┤ņŚ¼ņŻ╝ņŚłļŗż[12]. Random survival forest ĻĖ░ļ░ś ļ©ĖņŗĀļ¤¼ļŗØ ļ¬©ļŹĖĻ│╝ ĒĢ©Ļ╗ś, ņ×äņāüņØśņé¼ņŚÉĻ▓ī ņ╣£ņłÖĒĢ£ ņĀÉņłś ĻĖ░ļ░ś Ļ│äņé░Ēæ£ļź╝ ņĀ£ņŗ£ĒĢśņŚ¼ ņŚ¼ļ¤¼ ĒÖśĻ▓ĮņŚÉņä£ ĒÖ£ņÜ®ĒĢĀ ņłś ņ׳ļŖö ņ£äĒŚśļÅä ņśłņĖĪ ļ¬©ļŹĖņØä ņĀ£ņŗ£ĒĢśņŚ¼ ļ©ĖņŗĀļ¤¼ļŗØņØä ĒÖ£ņÜ®ĒĢ£ ņóŗņØĆ ņ×äņāüņŚ░ĻĄ¼ņØś Ēæ£ļ│ĖņØä ļ│┤ņŚ¼ņŻ╝Ļ│Ā ņ׳ļŗż.
ļ©ĖņŗĀļ¤¼ļŗØ ņĢīĻ│Āļ”¼ņ”śņØä ĒåĄĒĢ┤ ĻĖ░ņĪ┤ ņŚ░ĻĄ¼ļ░®ņŗØņ£╝ļĪ£ ĒāÉņāēĒĢśĻĖ░ ņ¢┤ļĀżņøĀļŹś ļŗżņ¢æĒĢ£ ņ╣śļŻīļ░śņØæĻĄ░ņØä Ļ░£ļ│äĒÖöĒĢśņŚ¼ ņśłņĖĪĒĢĀ ņłś ņ׳ļŖö Ļ░ĆļŖźņä▒ņØ┤ ņ׳ļŗż. ņŚ░ĻĄ¼ņ×ÉļōżņØĆ ACCORD ņŚ░ĻĄ¼ņ×ÉļŻīļź╝ Ļ▓ĆĒåĀĒĢśņŚ¼, ĻĖ░ņĪ┤ Ēæ£ņżĆ Ēśłļŗ╣Ļ┤Ćļ”¼ ļīĆļ╣ä ņ¦æņżæņĀüņØĖ Ēśłļŗ╣Ļ┤Ćļ”¼ļź╝ ņŗ£Ē¢ēĒĢśņśĆņØä ļĢī ļ│┤ņØ┤ļŖö ļŗżņ¢æĒĢ£ ļ░śņØæĻĄ░ņØä ņ×¼ļČäņäØĒĢśņśĆļŗż. ļ╣äļĪØ 2008ļģäļÅä ļ░£Ēæ£ļÉ£ ACCORD ņŚ░ĻĄ¼ņØś Ļ▓░ļĪĀņØĆ ņ¦æņżæĒśłļŗ╣Ļ┤Ćļ”¼Ļ░Ć Ēæ£ņżĆņ╣śļŻīļ▓ĢņŚÉ ļ╣äĒĢ┤ ņé¼ļ¦ØļźĀņØä ņ”ØĻ░Ćņŗ£Ēé©ļŗżļŖö Ļ▓āņØ┤ņŚłņ¦Ćļ¦ī, ļ©ĖņŗĀļ¤¼ļŗØ ņĢīĻ│Āļ”¼ņ”śņØä ĒåĄĒĢ£ ņé¼ĒøäļČäņäØņŚÉņä£ ņŚ░ĻĄ¼ņ×ÉļōżņØĆ ņ¦æņżæĒśłļŗ╣Ļ┤Ćļ”¼Ļ░Ć ņŗżņĀ£ ņé¼ļ¦ØļźĀ Ļ░£ņäĀņŚÉ ņ£ĀņØĄĒ¢łļŹś ļīĆņāüĻĄ░ņØä ņ░ŠņØä ņłś ņ׳ņŚłņ£╝ļ®░, ņāüļīĆņĀüņ£╝ļĪ£ ņé¼ļ¦ØļźĀ ņ”ØĻ░ĆņŚÉ ņśüĒ¢źņØä ņżĆ Ļ│Āņ£äĒŚśĻĄ░ņØĆ ņĀäņ▓┤ ļīĆņāüĻĄ░ ņżæņŚÉņä£ ņåīņłśņ×äņØä ņĀ£ņŗ£ĒĢśņŚ¼ ĻĖ░ņĪ┤ ņŚ░ĻĄ¼Ļ▓░Ļ│╝ļź╝ ņ×¼ĒĢ┤ņäØĒĢĀ ņłś ņ׳ļŖö Ļ░ĆļŖźņä▒ņØä ļ│┤ņŚ¼ņŻ╝ņŚłļŗż[13]. ņØ┤ ņŚ░ĻĄ¼ļŖö ĻĖ░ņĪ┤ ļīĆĻĘ£ļ¬© ļ¼┤ņ×æņ£äļīĆņĪ░ĻĄ░ ņŚ░ĻĄ¼ņØś Ļ▓░Ļ│╝Ļ░Ć ņŗĀņżæĒĢśĻ▓ī ĒĢ┤ņäØļÉĀ ĒĢäņÜöĻ░Ć ņ׳ņ£╝ļ®░, ļ©ĖņŗĀļ¤¼ļŗØņØä ĒÖ£ņÜ®ĒĢśņŚ¼ ļÅÖņØ╝ņ╣śļŻīļ▓ĢņŚÉ ļīĆĒĢ┤ ļŗżņ¢æĒĢ£ ņ╣śļŻīļ░śņØæņØä ļ│┤ņØ╝ ņłś ņ׳ļŖö ļ╣äĻĘĀņ¦łĒĢ£ ņ¦æļŗ©ņØä ņ░ŠņĢäļé┤Ļ│Ā ņØ┤ņŚÉ ļ¦×ņČś Ļ░£ļ│äĒÖöļÉ£ ņ╣śļŻīņĀü ņĀæĻĘ╝ņØä Ļ│ĀļĀżĒĢĀ ņłś ņ׳ņØīņØä ļ│┤ņŚ¼ņŻ╝ņŚłļŗż. ĒĢ£ ņŚ░ĻĄ¼ļŖö Ļ░ĢĒÖöĒĢÖņŖĄņØä ĒåĄĒĢ┤ ņĀ£1ĒśĢ ļŗ╣ļć©ļ│æ ĒÖśņ×ÉņŚÉņä£ ņÜ┤ļÅÖļ¤ē, ņŗØņØ┤ļ¤ē ļō▒ ļ│ĆĒÖöĒĢśļŖö ĒÖśĻ▓ĮņĪ░Ļ▒┤ņØä ļ░śņśüĒĢśņŚ¼ ņØĖņŖÉļ”░ ĒĢäņÜöļ¤ē Ļ▓░ņĀĢņØä ļÅäņÜĖ ņłś ņ׳ļŖö Ļ░ĆļŖźņä▒ņØä ņĀ£ņŗ£ĒĢśņśĆļŗż[5]. ņĢäņ¦ü ņä▒ļŖźņØ┤ ņČ®ļČäĒĢśņ¦ä ņĢŖņ¦Ćļ¦ī, ņØ┤ļ¤¼ĒĢ£ ņŚ░ĻĄ¼Ļ▓░Ļ│╝ļōżņØĆ ļ©ĖņŗĀļ¤¼ļŗØņØ┤ ņ╣śļŻīļ░śņØæ ņśłņĖĪ ļ░Å ņØ┤ļź╝ ĒåĄĒĢ£ Ļ░£ļ│äĒÖöļÉ£ ņĀæĻĘ╝ņØä Ļ░ĆļŖźĒĢśĻ▓ī ĒĢśļŖö ņżæņÜöĒĢ£ ļÅäĻĄ¼ļĪ£ ĒÖ£ņÜ®ļÉĀ Ļ░ĆļŖźņä▒ņØä ņŗ£ņé¼ĒĢ£ļŗż.
ļ©ĖņŗĀļ¤¼ļŗØ ņĢīĻ│Āļ”¼ņ”śņØĆ Ēśäņ×¼ ņŻ╝ļ¬®ņØä ļ░øĻ│Ā ņ׳ļŖö ļŗżņżæņśżļ»╣ņŖżļź╝ ĻĖ░ļ░śņ£╝ļĪ£ ĒĢ£ ņ×äņāü-ņżæĻ░£ņŚ░ĻĄ¼ņ×ÉļŻī ļČäņäØņŚÉņä£, ņłśļ¦ÄņØĆ ļ│Ćņłśļōż ņżæ ņŻ╝ņÜö ļ│Ćņłśļź╝ ņäĀĒāØĒĢśĻ│Ā ļ¬©ļŹĖņØä ĒÜ©Ļ│╝ņĀüņ£╝ļĪ£ ĻĄ¼ņČĢĒĢśĻĖ░ ņ£äĒĢ£ ņŻ╝ņÜöĒĢ£ ļÅäĻĄ¼ļĪ£ ĒÖ£ņÜ®ļÉĀ ņłś ņ׳ļŗż. Liu ļō▒[14]ņØĆ ņé¼ļ×īņŚÉ ņä£ ņÜ┤ļÅÖ Ēøä Ēśłļŗ╣ ļ│ĆĒÖö ļ░śņØæņä▒Ļ│╝ ņŚ░Ļ┤ĆļÉ£ ņŻ╝ņÜö ņןļé┤ļ»ĖņāØļ¼╝ĻĘĀņ┤Ø ļ░Å ļīĆņé¼ņ▓┤ļź╝ ĒāÉņāēĒĢśĻĖ░ ņ£äĒĢ┤ ļ©ĖņŗĀļ¤¼ļŗØ ņĀæĻĘ╝ļ▓ĢņØä ĒÖ£ņÜ®ĒĢśņśĆļŗż. ņÜ┤ļÅÖļ░śņØæĻĄ░Ļ│╝ ļ╣äļ░śņØæĻĄ░ņØĆ ņןļé┤ņäĖĻĘĀņ┤Ø ļ░Å ļīĆņé¼ņ▓┤ Ēī©Ēä┤ņŚÉ ņ׳ņ¢┤ ņ£ĀņØśĒĢ£ ņ░©ņØ┤ļź╝ ļ│┤ņśĆņ£╝ļ®░, ļČäļ│ĆņØ┤ņŗØņłĀņØä ĒåĄĒĢ┤ ņźÉņŚÉĻ▓ī ņÜ┤ļÅÖļ░śņØæĻĄ░ņØś ņןļé┤ņäĖĻĘĀņ┤Ø Ēī©Ēä┤ņØä ļ¦īļōżņ¢┤ ņŻ╝ņŚłņØä ļĢī ņÜ┤ļÅÖ ļ░Å ņØĖņŖÉļ”░ļ»╝Ļ░ÉļÅä Ļ░£ņäĀņŚÉ ĒÜ©Ļ│╝ļź╝ ļ│┤ņśĆļŗż. Random forest ņĢīĻ│Āļ”¼ņ”śņØĆ ņłśņ▓£ Ļ░£ņØś ņ×Āņ×¼ņĀü Ēøäļ│┤ ĒŖ╣ņä▒ļ│Ćņłśļōż Ļ░ĆņÜ┤ļŹ░ ļ░śņØæĻĄ░Ļ│╝ ļ╣äļ░śņØæĻĄ░ņØä Ļ░Ćņן ņל Ļ░Éļ│äĒĢśļŖö 29Ļ░£ņØś ĒŖ╣ņä▒ļ│Ćņłś(ņןļé┤ļ»ĖņāØļ¼╝ 14ņóģ, 15Ļ░£ ļīĆņé¼ņ▓┤) ņĪ░ĒĢ®ņØä ņ░ŠņĢśņ£╝ļ®░, ņØ┤ļ¤¼ĒĢ£ ĒŖ╣ņä▒ļ│ĆņłśņØś ņĪ░ĒĢ®ņØĆ ņÜ┤ļÅÖļ░śņØæĻĄ░Ļ│╝ ļ╣äļ░śņØæĻĄ░ņØä ņśłņĖĪĒĢśĻ│Ā ņØ┤ņŚÉ ļö░ļźĖ ņ╣śļŻīņĀäļץņØä ņäĖņÜĖ ņłś ņ׳ļŖö Ļ░£ļ│äĒÖöļÉ£ ņĀæĻĘ╝ņØä ņ£äĒĢ£ ļ░öņØ┤ņśżļ¦łņ╗żļĪ£ ĻĖ░ļŖźĒĢĀ Ļ░ĆļŖźņä▒ņØ┤ ņ׳ļŗż.
ĻĄŁļé┤ ļ©ĖņŗĀļ¤¼ļŗØ ĒÖ£ņÜ® ņØśĒĢÖņŚ░ĻĄ¼ļŖö ļöźļ¤¼ļŗØ ĻĖ░ļ▓ĢņØś ļ░£ņĀäĻ│╝ ĒĢ©Ļ╗ś ņŻ╝ļĪ£ ņśüņāüņØśĒĢÖ ļČäņĢ╝ņŚÉņä£ ĒÖ£ļ░£ĒĢśĻ▓ī ņØ┤ļŻ©ņ¢┤ņĪīĻ│Ā, ļŗ╣ļć©ļ│æ ļ░Å ļé┤ļČäļ╣äņ¦łĒÖś ņ×äņāüņŚ░ĻĄ¼ņŚÉņä£ļÅä ņĄ£ĻĘ╝ ņĀüĻĘ╣ņĀüņ£╝ļĪ£ ļ©ĖņŗĀļ¤¼ļŗØņØä ĒÖ£ņÜ®ĒĢśļĀżļŖö ņŚ░ĻĄ¼ņ×ÉļōżņØ┤ ļŖśņ¢┤ļéśĻ│Ā ņ׳ļŗż. ļ│æņøÉņ×ÉļŻī ĻĖ░ļ░śņ£╝ļĪ£ 5ļģä ļé┤ ļŗ╣ļć©ļ░£ņāØļźĀņØä ņśłņĖĪĒĢśļŖö ļ¬©ļŹĖņØä ņĀ£ņŗ£ĒĢ£ ņŚ░ĻĄ¼ņŚÉņä£ļŖö, ņĀäņ×ÉņØśļ¼┤ĻĖ░ļĪØ ĻĖ░ļ░ś 28Ļ░£ ļ│Ćņłśļź╝ ņČöņČ£ĒĢśņŚ¼ ļ©ĖņŗĀļ¤¼ļŗØ ļ¬©ļŹĖņØä ļ¦īļōżĻ│Ā ļŗ╣ļć©ļ│æ ļ░£ņāØ ņŚ¼ļČĆļź╝ ņśłņĖĪĒĢśņśĆļŗż. ļŗżļ¦ī ņśłņĖĪļĀźņŚÉ ņ׳ņ¢┤ņä£ ĻĖ░ņĪ┤ ņśłņĖĪļ¬©ļŹĖņØä ņāüĒÜīĒĢśļŖö ņä▒ļŖźņØä ļ│┤ņŚ¼ņŻ╝ņ¦ĆļŖö ļ¬╗ĒĢśņŚ¼, ĻĖ░ņĪ┤ ļ│æņøÉņ×ÉļŻīņŚÉņä£ ņ¢╗ņØä ņłś ņ׳ļŖö ņśłņĖĪļĀźņØś ĒĢ£Ļ│äļź╝ ļäśĻĖ░ ņ£äĒĢ┤ņä£ļŖö ņČöĒøä ĒÖśņ×ÉņØś ņāØĒÖ£Ēī©Ēä┤ņØ┤ļéś ņŗØņŖĄĻ┤Ć, ņÜ┤ļÅÖ ļō▒ ļ│æņøÉ ļ░¢ņŚÉņä£ ņłśņ¦æļÉĀ ņłś ņ׳ļŖö ĒŖ╣ņä▒ļ│ĆņłśņØś ĒÖ£ņÜ®ņØ┤ ņżæņÜöĒĢĀ Ļ▓āņ£╝ļĪ£ ļ│┤ņØĖļŗż[16]. ņĄ£ĻĘ╝ ĒÖ£ņÜ®ņØ┤ ņ”ØĻ░ĆĒĢśĻ│Ā ņ׳ļŖö ņŚ░ņåŹĒśłļŗ╣ņĖĪņĀĢĻĖ░(continuous glucose monitoring, CGM)ņŚÉņä£ ņ¢╗ņØä ņłś ņ׳ļŖö Ēśłļŗ╣ ļ│ĆĒÖö ņŗ£Ļ│äņŚ┤ ļŹ░ņØ┤Ēä░ļŖö ļ©ĖņŗĀļ¤¼ļŗØņØä ĒÖ£ņÜ®ĒĢśĻĖ░ņŚÉ ņĀüņĀłĒĢ£ ņ×ÉļŻīņØ┤ļŗż. ĒĢ£ ĻĄŁļé┤ ņŚ░ĻĄ¼ņŚÉņä£, ņŚ░ĻĄ¼ņ¦äņØĆ 30ļČä ņØ┤ļé┤ ņĀĆĒśłļŗ╣ņØ┤ ļ░£ņāØĒĢĀ Ļ▓āņØä ņśłņĖĪĒĢśļŖö ļ¬©ļŹĖņØä ĻĄ¼ņČĢĒĢśņśĆļŗż. Random forest ļ¬©ļŹĖņØĆ area under the receiver-operating characteristics curve 0.966, ļ»╝Ļ░ÉļÅä 89.6%, ĒŖ╣ņØ┤ļÅä 91.3%ļĪ£ ņóŗņØĆ ņśłņĖĪ ļŖźļĀźņØä ļ│┤ņŚ¼ ņČöĒøä CGM ļ░Å ņØĖĻ│ĄņĘīņן Ļ░£ļ░£, Ļ│ĀļÅäĒÖöņŚÉ ļÅäņøĆņØä ņżä Ļ░ĆļŖźņä▒ņØ┤ ņĀ£ņŗ£ĒĢśņśĆļŗż[17].
ļ©ĖņŗĀļ¤¼ļŗØņØś ĒÖ£ņÜ®ņØĆ ļŹ░ņØ┤Ēä░ ĒÖ£ņÜ®, ĒĢ┤ņäØņŚÉ ņ׳ņ¢┤ ņāłļĪ£ņÜ┤ Ļ░ĆļŖźņä▒ņØä ņĀ£ņŗ£ĒĢśļ®░ ļŗ╣ļć©ļ│æ ļ░Å ļīĆņé¼ņ¦łĒÖś ņŚ░ĻĄ¼ņŚÉ ļÅäņøĆņØä ņżä Ļ░ĆļŖźņä▒ņØ┤ ņ׳ņ¦Ćļ¦ī, Ēśäņ×¼ ņØśļŻīļŹ░ņØ┤Ēä░ņØś ļ│Ąņ×Īņä▒, ļČĆņĀĢĒÖĢņä▒, ņ×Āņ×¼ņĀü ņśżļźśļź╝ Ļ│ĀļĀżĒĢĀ ļĢī ļīĆĻĘ£ļ¬© ņ×ÉļŻīņØś ĒÖĢļ│┤ņÖĆ ļ©ĖņŗĀļ¤¼ļŗØņØś ņĀüņÜ®ņØĆ ņŗĀņżæĒĢ£ ņĀæĻĘ╝ņØ┤ ĒĢäņÜöĒĢśļŗż[18]. ņĀĢĒÖĢĒĢ£ ļ¼ĖņĀ£ ņĀĢņØś Ēś╣ņØĆ Ļ░Ćņäż ņäżņĀĢ, ļŹ░ņØ┤Ēä░ ņ¦ł(quality)ņŚÉ ļīĆĒĢ£ ņ¦ĆņåŹņĀüņØĖ Ļ┤Ćņŗ¼, ņ×äņāüĒśäņן ļ░Å ļ»ĖņČ®ņĪ▒ņłśņÜöņŚÉ ĻĖ░ļ░śĒĢ£ ņŚ░ĻĄ¼ļööņ×ÉņØĖ ļ░Å ļ©ĖņŗĀļ¤¼ļŗØ ņĀäļ¼ĖĻ░Ć ņ╗żļ«żļŗłĒŗ░ņÖĆņØś ņĀüĻĘ╣ņĀüņØ┤Ļ│Ā Ēł¼ļ¬ģĒĢ£ ĒśæļĀźņØ┤ ļ©ĖņŗĀļ¤¼ļŗØņØä ĒåĄĒĢ£ ļŗ╣ļć©ļ│æ ļ░Å ļīĆņé¼ņ¦łĒÖś ņŚ░ĻĄ¼ļź╝ ņä▒Ļ│ĄņĀüņ£╝ļĪ£ ņŗ£Ē¢ēĒĢĀ ņłś ņ׳ļŖö ņżæņÜöĒĢ£ ņÜöņåīĻ░Ć ļÉĀ ņłś ņ׳ļŗż[19]. ļ©ĖņŗĀļ¤¼ļŗØņØä ļŗ╣ļć©ļ│æĻ│╝ ļé┤ļČäļ╣äņ¦łĒÖś ņŚ░ĻĄ¼ņŚÉ ĒÜ©Ļ│╝ņĀüņ£╝ļĪ£ ņĀüņÜ®ĒĢśņŚ¼ ņ×äņāüņĀüņ£╝ļĪ£ ņ£ĀņÜ®ĒĢ£ Ļ▓░Ļ│╝ļź╝ ņØ┤ļüīņ¢┤ļé┤ĻĖ░ ņ£äĒĢ┤ņä£ļŖö, ļīĆĻĘ£ļ¬©, Ļ│ĀĒÆłņ¦łņØś ņØśļŻīļŹ░ņØ┤Ēä░ņØś ĻĄ¼ņČĢ ļ░Å ņĀæĻĘ╝ņä▒ ĒÖĢļ│┤Ļ░Ć ņżæņÜöĒĢśļŗż. ņØ┤ļź╝ ņ£äĒĢśņŚ¼ ļ│æņøÉņ×ÉļŻī, ņ¦ĆņŚŁņé¼ĒÜīņĮöĒśĖĒŖĖ, ĒÖśņ×É ļĀłņ¦ĆņŖżĒŖĖļ”¼, Ļ▒┤Ļ░Ģļ│┤ĒŚś ņ▓ŁĻĄ¼ņ×ÉļŻī, Ļ░£ņØĖ ļØ╝ņØ┤ĒöäļĪ£ĻĘĖ ļō▒ ņŚ¼ļ¤¼ ņ×ÉļŻīļōżņØ┤ ņØĄļ¬ģņä▒ņØä ņ£Āņ¦ĆĒĢ£ ņ▒ä ņŚ░Ļ│äļÉ£ ļŗżļ®┤ļŹ░ņØ┤Ēä░ ĻĄ¼ņČĢ, ņĀĢņĀ£ ļ░Å Ļ│ĄĒåĄņ×ÉļŻīļ¬©ļŹĖ ļō▒ņØä ņØ┤ņÜ®ĒĢ£ ĒÜ©ņ£©ņĀüņØĖ ļŗżĻĖ░Ļ┤Ć ņŚ░ĻĄ¼ ļ¬©ļŹĖ ņłśļ”ĮņØ┤ ĒĢäņÜöĒĢśļŗż. ņĀÉņ░© ņ”ØĻ░ĆĒĢśļŖö ļ©ĖņŗĀļ¤¼ļŗØ ĻĖ░ļ░ś ņŚ░ĻĄ¼Ļ▓░Ļ│╝ļōżņØä ņĀĢĒÖĢĒĢśĻ▓ī ņØ┤ĒĢ┤, ļ╣äĒīÉņĀü ĒÅēĻ░Ć Ēøä ņ×äņāüņŚÉ ļÅäņ×ģĒĢśņŚ¼ ņŗżņĀ£ ĒÖśņ×ÉļōżņŚÉĻ▓ī ļÅäņøĆņØä ņżä ņłś ņ׳ļÅäļĪØ ņ¦äļŻīĒśäņןņØä ļ│ĆĒÖöņŗ£ĒéżĻĖ░ ņ£äĒĢ┤ņä£ļŖö, ņĀäļ¼Ėņ×äņāüĻ▓ĮĒŚśņØä ļ│┤ņ£ĀĒĢśĻ│Ā ņ׳ņ£╝ļ®░ ņ×äņāüĒśäņןņØś ļ»ĖņČ®ņĪ▒ņłśņÜöļź╝ ņל ņØĖņ¦ĆĒĢśĻ│Ā ņ׳ļŖö ļŗ╣ļć©ļ│æ ļ░Å ļīĆņé¼ņ¦łĒÖś ļé┤ļČäļ╣ä ņ×äņāüņØśņé¼ņØś ņŚŁĒĢĀņØ┤ ĒĢäņłśņĀüņØ┤ļŗż.
Table┬Ā1.
Machine learning algorithms
| Types of learning | Supervised learning | Semi-supervised learning | Reinforcement learning | Unsupervised learning |
|---|---|---|---|---|
| Concept | Learning a function that best approximates new input to the desired output based on a given relationship between the input and labeled output from the labeled dataset | A mixed approach of supervised and unsupervised learning applicable to a small amount of labeled data and a large amount of unlabeled data | Learning by maximizing the reward function based on the responses yielded by various actions to achieve arbitrary goals in a given unstructured or unknown environment | Finding structures or patterns in an unlabeled dataset |
| Common tasks | Regression, classification | Regression, classification | Taking actions to maximize the reward | Clustering, dimensionality reduction |
| Estimators | Naïve Bayesian, decision tree, support vector machine (SVM), neural network, logistic/ridge/ linear regression, elastic net, etc. | Generative model, semi-supervised SVM, etc. | Q-learning, policy gradient, actor-critic, etc. | K-means, density-based spatial clustering of applications with noise, auto-encoders, deep Boltzmann machine, principal component analysis, locally linear embedding, etc. |
| Examples | Prediction of gestational diabetes according to biochemical test results based on simple features extracted from an electronic health records database [4] | The DeepHeart algorithm, which provides cardiovascular risk scores based on heart rate monitoring from popular wearable devices (Fitbit, Apple Watch, etc.) [6] | Determining the optimal insulin dose in patients with type 1 diabetes based on activity, hemoglobin A1c level, alcohol consumption status, and the previous insulin dose [5] | Identifying novel clusters or biomarkers based on various features collected by an unbiased multimodal approach, which finds differences in risks for certain diseases compared to other groups [7] |
Table┬Ā2.
| Metrics | Concepts and equations |
|---|---|
| Accuracy | ŌĆó (TP + TN) / (TP + FP +FN + TN) |
| Precision (= positive predictive value) | ŌĆó TP/(TP + FP) |
| Recall (= sensitivity, true positive rate) | ŌĆó TP/(TP + FN) |
| F1-score | ŌĆó The harmonic mean of precision and recall |
| ŌĆó 2 ├Ś (recall ├Ś precision) / (recall + precision) | |
| AUROC | ŌĆó The area under the receiver operating characteristic curve (plotting TPR against FPR) |
| ŌĆó Higher AUROC, close to 1 = better classifier | |
| AUPRC | ŌĆó The area under the precision-recall curve (plotting precision against recall) |
| ŌĆó Higher AUPRC, close to 1 = better classifier | |
| ŌĆó May have an advantage over AUROC when comparing the performance of models in an imbalanced dataset [10] |
Table┬Ā3.
Summary of recent studies related to machine learning applications in diabetes research
| Task | Study (disease field) | Study subjects | Design and method | Key finding and limitation |
|---|---|---|---|---|
| Screening and diagnosis | Artzi et al., 2020 [4] | - Retrospective nationwide electronic health record data of 588,622 pregnancies from 368,351 women between 2010 to 2017 in Israel including data of demographics, anthropometrics, laboratory tests, diagnoses, and pharmaceuticals | - Aim: to establish an ML model to improve the prediction of gestational diabetes based on electronic health record vs. a conventional screening tool | Key implications |
| - ML was useful in developing a simple nine-question model in self-reportable format from the large electronic health record dataset, which outperformed the current standard screening tool (AUROC 0.80 vs. 0.68). | ||||
| - Internal validation set (n = 137,220; with geotemporal difference) | - Reference labels: gestational diabetes diagnosis by a twostep approach (glucose challenge test and oral glucose tolerance test at 24~28 weeks of gestation) | - May facilitate early-stage interventions for women at high risk for gestational diabetes | ||
| - Comparator: National Institute of Health sevenitem questionnaire | - May aid construction of a selective, cost-effective screening approach according to predicted gestational diabetes risk instead of the current universal screening approach Limitations | |||
| - Inherent bias from retrospective electronic health record data review | ||||
| - Methods: supervised learning; gradient boosting model | - Performance might be different when based on actual self-reported surveys. | |||
| Perakakis et al., 2019 [11] | - Serum samples of 49 healthy subjects and 31 patients with biopsyproven NAFLD | - Aim: to train models for the non-invasive diagnosis of NASH and liver fibrosis based on circulating lipids, glycans, fatty acids identified by liquid Chromatography with tandem mass spectrometry LC-MS/MS and biochemical parameters | Key implications | |
| - Internal validation with three-fold crossvalidation | - The ML model including 20 features consisted of lipidomics, glycans, and adiponectin yielded high accuracy up to 90% in discriminating healthy individuals from patients with NAFLD and NASH. | |||
| - Reference label: biopsyproven NAFLD | - May provide a low-risk cost-effective, non-invasive alternative method to liver biopsy. | |||
| - Comparator: not applicable - Methods: supervised learning; one-vs-rest nonlinear support vector machine models with recursive feature elimination | Limitations | |||
| - Validation cohort was not available | ||||
| - Needs to be further validated in a different population. | ||||
| Risk prediction | Segar et al., 2019 [12] | - 8,756 Patients without heart failure at baseline from the ACCORD trial dataset (50% training set; 50% internal validation set; conducted between 1999 to 2009) | - Aim: to develop an ML model to predict incident heart failure among patients with type 2 diabetes | Key implications |
| - The ML-based models showed modest performance in prediction for incident heart failure among patients with type 2 diabetes in the external validation set (C-index 0.70 to 0.74). | ||||
| - External validation set: 10,819 participants without prevalent heart failure from the ALLHAT trial | - Reference label: incident hospitalization or death due to heart failure (captured and adjudicated by two independent reviewer physicians during the trial) | - Each 1-unit increment in the WATCHDM score was associated with a 24% higher relative risk of heart failure within 5 years | ||
| - Strength of analyzing a large number of participants from a well-phenotyped clinical trial population Limitations | ||||
| - Comparator: not applicable | - Discrimination for heart failure with preserved ejection fraction was relatively low in the subgroup analysis. | |||
| - Methods: supervised learning; random survival forest-based model | - Temporal changes of heart failure biomarkers and medications could not be reflected in the model. | |||
| - Need to validate the model in lowerrisk cohorts of individuals with type 2 diabetes | ||||
| Basu et al., 2018 [13] | - 10,251 ACCORD trial participants aged 40 to 79 years with type 2 diabetes, HbA1c 7.5% or higher, or cardiovascular diseases or risk factors, those who randomized to target HbA1c < 6.0% (intensive) vs. 7.0% to | - Aim: to identify subgroups with a heterogeneous treatment effect in response to intensive glycemic therapy | Key implications | |
| - Compared to 3.7% increased mortality by intensive vs. standard therapy in group 4, group 1 showed a 2.3% mortality reduction in the intensive therapy group (95% CI, -0.2% to 4.5%), which made the obvious contrast with the main result from the study. | ||||
| -7.9% (standard group) | - Reference label: treatment effect defined as the absolute difference in the all-cause mortality rate between the intensive and standard therapy groups | - Identified characteristics of patients who may have benefited from intensive glycemic therapy (younger individuals with relatively low hemoglycosylation index) | ||
| - Offered an example to find clinically meaningful subgroups with heterogeneous treatment effects using data from randomized trials. | ||||
| - Comparator: not applicable | Limitations | |||
| - Methods: supervised learning; gradient forest analysis | - Post hoc analysis of a single trial that was conducted before the development of recent diabetes medications with cardiovascular benefits | |||
| Oroojeni et al., 2019 [5] | - Medical records of 87 patients with type 1 diabetes from Mass General Hospital; data for each patientŌĆÖs visits over a 10-year period (training set) between 2003 to 2013; HbA1c, body mass index, activity level, alcohol usage status, insulin (Lantus) dose | - Aim: to explore an effective reinforcement learning framework for determining the optimal long-acting insulin dose for patients with type 1 diabetes | Key implications | |
| - The physician-prescribed insulin dose was within the dosing interval recommended by the Q-learning algorithm in 88% of test cases. | ||||
| - External validation with 60 cases | - Reference label: physicianprescribed insulin dose | - A proof-of-concept study to provide clinical decision support for determining insulin dose in patients with type 1 diabetes, by applying reinforcement learning algorithm | ||
| Limitations | ||||
| - Comparator: not applicable | - Limited by omitting lifestyle information regarding diet, stress, and medication adherence | |||
| - Methods: reinforcement learning; Q-learning with reward function set from HbA1c status at the visit and change of HbA1c from the past visit | - A relatively small training set | |||
| - Only one type of insulin (Lantus) was examined in the model | ||||
| Translational research | Liu et al., 2020 [14] | - 20 Drug-naive individuals with prediabetes (discovery cohort) | - Aim: to find an ML model for predicting exercise responsiveness determined from exercise-induced alterations in the gut microbiota | Key implications |
| - Determined exercise responders and nonresponders after 12-week high-intensity exercise training | - The ML model identified 14 microbiome species and 15 metabolites from human feces were able to predict exercise responsiveness (AUROC 0.75 in the validation set). | |||
| - Reference label: responders defined as a decrease in the homeostatic model assessment of insulin resistance greater than two-fold technical error | - Provide an example of applying ML principles to human-tomice translational study based on microbiome dataset | |||
| - Collected pre- and postexercise period feces to analyze gut microbiota profile | Limitations | |||
| - Comparator: not applicable | - Relatively small sample size | |||
| - Internal validation with 10-fold cross-validation | - Methods: supervised learning; random forest model | - Limited to Chinese males only | ||
| - Need further validation in different population set |
REFERENCES
2. McCarthy J. What is artificial intelligence? Available from: http://www-formal.stanford.edu/jmc/whatisai.html (Accessed on 9th June, 2020).
3. Beaulieu-Jones B, Finlayson SG, Chivers C, Chen I, McDermott M, Kandola J, et al. Trends and focus of machine learning applications for health research. JAMA Netw Open 2019;2:e1914051.



4. Artzi NS, Shilo S, Hadar E, Rossman H, Barbash-Hazan S, Ben-Haroush A, et al. Prediction of gestational diabetes based on nationwide electronic health records. Nat Med 2020;26:71-6.


5. Oroojeni Mohammad Javad M, Agboola SO, Jethwani K, Zeid A, Kamarthi S. A reinforcement learning-based method for management of type 1 diabetes: exploratory study. JMIR Diabetes 2019;4:e12905.



6. Ballinger B, Hsieh J, Singh A, Sohoni N, Wang J, Tison GH, et al. DeepHeart: semi-super vised sequence learning for cardiovascular risk prediction. CoRR 2018;arXiv:1802.02511.

7. Shomorony I, Cirulli ET, Huang L, Napier LA, Heister RR, Hicks M, et al. An unsupervised learning approach to identify novel signatures of health and disease from multimodal data. Genome Med 2020;12:7.



8. Dinga R, Penninx BWJH, Veltman DJ, Schmaal L, Marquand AF. Beyond accuracy: measures for assessing machine learning models, pitfalls and guidelines. bioRxiv 2019;743138.

9. Handelman GS, Kok HK, Chandra RV, Razavi AH, Huang S, Brooks M, et al. Peering into the black box of artificial intelligence: evaluation metrics of machine learning methods. AJR Am J Roentgenol 2019;212:38-43.


10. Saito T, Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One 2015;10:e0118432.



11. Perakakis N, Polyzos SA, Yazdani A, Sala-Vila A, Kountouras J, Anastasilakis AD, et al. Non-invasive diagnosis of non-alcoholic steatohepatitis and fibrosis with the use of omics and supervised learning: a proof of concept study. Metabolism 2019;101:154005.


12. Segar MW, Vaduganathan M, Patel KV, McGuire DK, Butler J, Fonarow GC, et al. Machine learning to predict the risk of incident heart failure hospitalization among patients with diabetes: the WATCH-DM risk score. Diabetes Care 2019;42:2298-306.



13. Basu S, Raghavan S, Wexler DJ, Berkowitz SA. Characteristics associated with decreased or increased mortality risk from glycemic therapy among patients with type 2 diabetes and high cardiovascular risk: machine learning analysis of the ACCORD trial. Diabetes Care 2018;41:604-12.


14. Liu Y, Wang Y, Ni Y, Cheung CKY, Lam KSL, Wang Y, et al. Gut microbiome fermentation determines the efficacy of exercise for diabetes prevention. Cell Metab 2020;31:77-91.e5.


15. Katsiki N, Gastaldelli A, Mikhailidis DP. Predictive models with the use of omics and supervised machine learning to diagnose non-alcoholic fatty liver disease: a "non-invasive alternative" to liver biopsy? Metabolism 2019;101:154010.


16. Choi BG, Rha SW, Kim SW, Kang JH, Park JY, Noh YK. Machine learning for the prediction of new-onset diabetes mellitus during 5-year follow-up in non-diabetic patients with cardiovascular risks. Yonsei Med J 2019;60:191-9.



17. Seo W, Lee YB, Lee S, Jin SM, Park SM. A machine-learning approach to predict postprandial hypoglycemia. BMC Med Inform Decis Mak 2019;19:210.



-
METRICS

-
- 1 Crossref
- 0 Scopus
- 3,473 View
- 67 Download
- Related articles
-
A Case on the Application of Diabetes Conversation Map2009 December;10(4)
Diabetes and Endocrine Disease.2017 September;18(3)
Connections between Diabetes and Cognitive Dysfunction.2019 June;20(2)



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print